Abstract
WIVACE is an Italian word meaning "Lively" as it shows in google translate. WIVACE is an abbreviation for Artificial Life and Evolutionary Computation: 17th Italian workshop, WIVACE 2023, Venice, Italy, September 6-8, 2023 at the University of Venice. This report provides a detailed summary, media, and author details from the various presentations, keynotes and talks. From our "The Living Technology Lab", (me, Prof Stefano and Kristine Heiney, PhD) we presented two papers at WIVACE 2023. There were many relevant talks and presentations from different professors, postdocs and PhDs at the conference of which some of it are summarised in this text. The conference Schedule plan for the program has more details about the papers presented and keynotes. We also met some notable researchers and recognised faces in the research areas including but not limited to Emergent Complexity, ALife, Computational Biology, Evo-Devo and Evolutionary Robotics, community starting from Prof. Andrea Roli, Prof. Wolfgang Banzhaf, Joana C. Xavier, Ph.D., Prof. JJ Merelo, Prof. Roberto Serra, Prof. Paolo Pellizzari, Vassilis Papadopoulos, Paolo Baldini, Michiel Stock, Michiel Stock, Clément Hongler, Marco Tuccio and many other notable researchers from the community. But we also missed Eric Medvet and Bert Chan. Finally I would like to thank Monica Kristiansen Holone for all support from Østfold University College, Norway and also OsloMet University, Norway. The Conference venue was: Aula Mario Baratto, Dorsoduro 3246 - 30123 Venice and European Centre for Living Technology (ECLT) Ca' Bottacin, Dorsoduro 3911, Calle Crosera, 30123 Venice, Italy. Most probably the WIVACE 2024 is expected to being held in NAXYS, UNIVERSITY OF NAMUR UNamur
Day 1 - 6th September 2023
There were multiple presentations and one keynote from day 1. Particularly there were two presentations and one keynote by Dr. Joana I would be presenting as summary for day 1. Following are the breif discussions from the respective presentations.
M1 Morning session 1: (25 min) Marco Villani, Matteo Balugani and Roberto Serra with title as “The properties of pseudo-attractors in random Boolean networks”
To start with in sequence I found this paper: "Marco Villani, Matteo Balugani and Roberto Serra: The properties of pseudo-attractors in random Boolean networks" at kind of a preprint.
I found this extended abstract quite similar to the work of coarse graining. If you remember our coarse graining in CA (proposed with MNCA) the proposed idea in this paper is quite similar to that, but for Random Boolean Networks. They introduce "Pseudo-attractors" to capture the average behavior of genes or components within the network over time, even if the system gets chaotic.
The idea is they hypothesize and later show that genes in single real-world cells are often chaotic; however, they are interdependent. At the same time, while in the simulation of the RBN, they try to capture the same notion using pseudo-attractors.
To define pseudo-attractors, they calculate the average state of each node (gene) across different time steps within an attractor. "If the average state of a node is mostly 1 (on) during an attractor, it's considered to be part of the pseudo-attractor," something like frequency, don't you think so?
Reducing or finding the periodic attractors from a chaotic one is something they emphasize a lot and then choose real-world data to compare and analyze gene expression or other cellular activities.
What are your thoughts? I think averaging or calculating the number of 1s is something related to frequency and then utilizing the "coarse graining" or they call it as averaging to identify pseudo-attractors.
Title: Pseudo-attractors in Random Boolean Network Models and Single-Cell Data
Abstract: This is a summary of a research paper that introduces two new ideas in a field called Random Boolean Networks, which are used to understand complex systems. These new ideas are called "pseudo-attractors" and the "common sea." The paper explains how these ideas can be used to understand how genes work in individual cells by analyzing their activity.
Introduction: Imagine you want to understand how a complex system works, like how genes in our cells interact. Scientists use models to help them understand complex things. One of these models is called a Random Boolean Network (RBN). It's like a simplified version of how genes in our cells turn on and off. The RBN model can represent different behaviors, like when things are very ordered or when they seem random. This model can also help us understand how genes work, even though it's not exactly like real cells.
In this RBN model, each gene's behavior is influenced by other genes and their rules for turning on and off. The model works in steps, where each step is like a moment in time. At each step, genes can change their state based on their rules and the states of other genes. This process is called "updating." However, in this paper, they're mostly focusing on a version where all the genes change at the same time, which might not be exactly how it happens in real cells.
Pseudo-attractors and Common Sea: Now, imagine you're watching how genes change over time. You might notice some patterns that genes follow. These patterns are called "attractors." Attractors can be stable states (like when genes settle down to a specific pattern) or cycles (like when genes keep repeating a pattern). But here's the tricky part: the patterns we see can change a lot based on how we're looking at them. It's like seeing different things depending on how we look at them.
So, in this paper, they introduce a new idea called "pseudo-attractors." These are like patterns that genes tend to follow on average, even if things seem a bit chaotic. To make it simple, they're finding a way to look at the average behavior of genes even if things seem to be changing a lot.
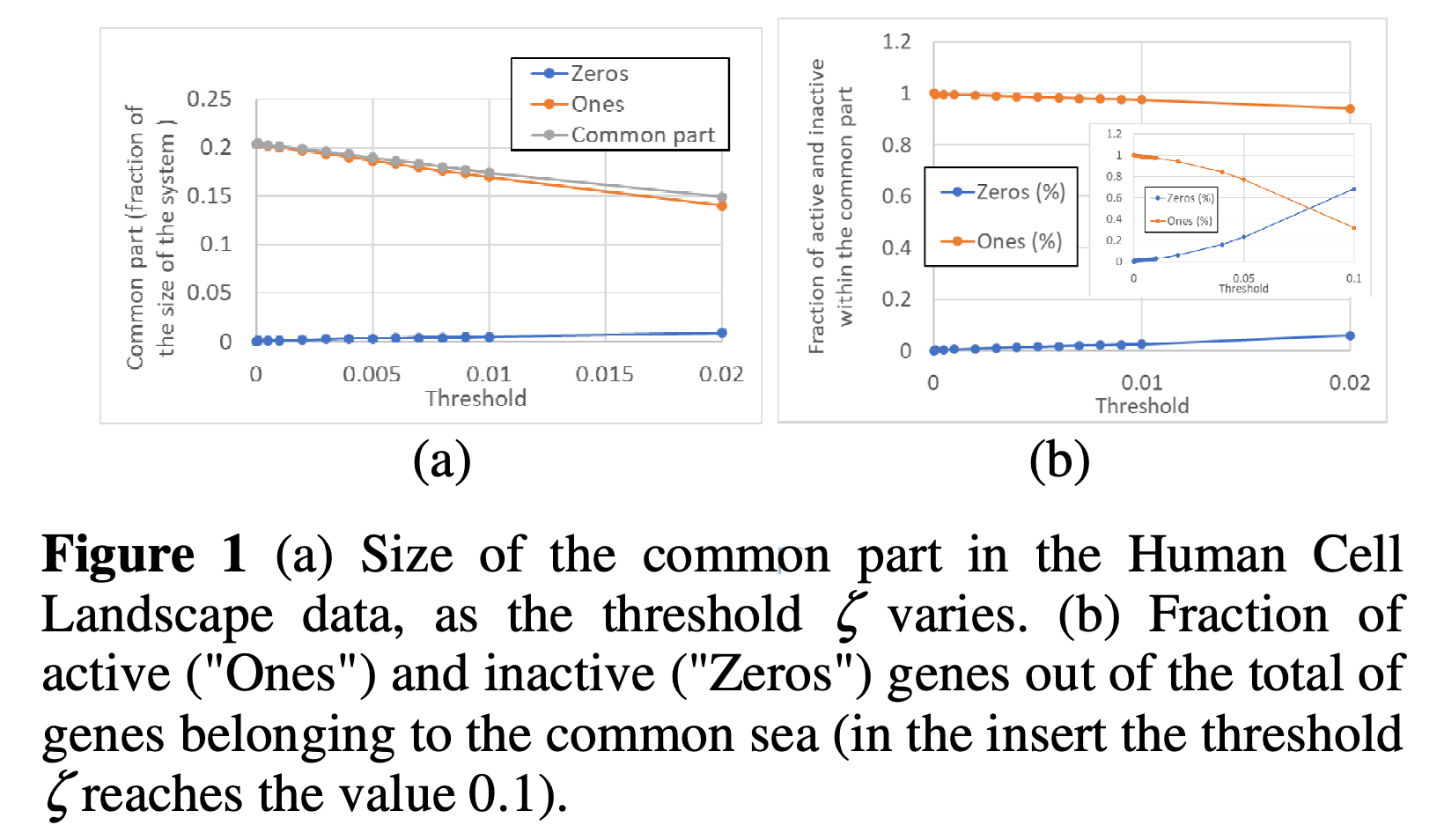
Then, they talk about the "common sea." Imagine you're comparing these patterns for different cells. Some genes might behave in similar ways in different cells. They call this the "common sea." It's like finding the parts that are the same in different cells. The rest of the genes are in the "specific part," which are the parts that are different between cells.
Applying to Real Data: Now, let's switch from the model to real cells. Scientists can measure how genes are behaving in single cells. But this data can be messy and hard to understand. The paper suggests using the ideas of pseudo-attractors and the common sea to make sense of this data.
For example, they took real data from cells and turned it into simpler patterns using pseudo-attractors. They found that some genes tend to behave similarly across different types of cells. This is like finding the common sea in real cells.
Conclusion: This paper introduces two new concepts, pseudo-attractors and the common sea, in a model called Random Boolean Networks. These concepts help us understand how genes might work in individual cells. They show that these ideas can also be useful for understanding real data from cells. This kind of work could help us better understand how genes behave in complex systems like our bodies.
A pseudo-attractor is a concept introduced in the context of Random Boolean Networks (RBNs) to capture the average behavior of genes or components within a network over time, even if the system's dynamics appear to be somewhat chaotic or variable.
-
Random Boolean Networks (RBNs):
An RBN is a mathematical model used to study complex systems, often applied to gene regulatory networks.
In an RBN, each "node" represents a gene or a component, and these nodes can be either in an "on" state (1) or an "off" state (0).
Nodes are connected in a network, and they influence each other's states based on specific rules or functions.
-
Attractors in RBNs:
In RBNs, "attractors" are patterns or states that the nodes tend to settle into over time. These can be stable points (where the system doesn't change) or cycles (where the system repeats a pattern).
-
Pseudo-attractors:
In real-world biological systems, such as gene expression in cells, the behavior of genes is influenced by various factors and can be quite complex and noisy.
Pseudo-attractors are introduced to capture the overall or average behavior of genes even if the system's dynamics seem chaotic or noisy.
Imagine if you watch a system evolving over time and it seems like it's changing a lot, but if you step back and look at it from a broader perspective, you might notice some patterns that repeat on average. These repeating patterns are like pseudo-attractors.
-
Calculating Pseudo-Attractors:
To define pseudo-attractors, researchers calculate the average state of each node (gene) across different time steps within an attractor.
If the average state of a node is mostly 1 (on) during an attractor, it's considered to be part of the pseudo-attractor.
This helps capture the genes' typical behavior during attractors, even if they might oscillate or change during individual steps.
-
Importance and Use:
Pseudo-attractors provide a way to understand the overall behavior of genes in a more stable manner, even if the system's dynamics seem complex.
They offer a simplified view of gene behavior that can be useful for interpreting experimental data or making connections between the RBN model and real-world biological systems.
Pseudo-attractors help bridge the gap between the abstract RBN model and the complexity of gene expression in actual cells.
In essence, a pseudo-attractor is like an "average behavior pattern" that helps us see the underlying order within the seemingly chaotic dynamics of a complex system like gene regulation.
M2 Morning session 2: (25 min) Prof. JJ Merelo and M. Cristina Molinari The complex networks of the Republic of Venice
The complex network of the Republic of Venice, presented at #WIVACE2023 in Venice by @jjmerelo wearing the outfit of Venezia football club 👍 pic.twitter.com/kM5SVCx64D
— Stefano Nichele (@stenichele) September 6, 2023
Read more relevant work at Molinari, M. C. (2023). Much ado about nothing: voting in sixteenth-century Republic of Genoa. Constitutional Political Economy, 1-22.
Dr. Joana C. Xavier Keynote Speech: Autocatalysis and the origins of life on Earth
Dr. Joana presented a presentation on very interetsting topic "Autocatalysis and the origins of life on Earth". It was a great presentation. Easy to understand while keeping the core to biology and sciences. This section provides detailed summary of the keynote of Dr. Joana. Before we jump to the summary, I would like to share some youtube videos and references which I watched as well to provide this summary. These youtube links are podcasts of Dr Joana where she explins Origin of Life on Earth, Physics of Emergent Behaviour and Intelligent Design in Emerging Complexity. For the slides snapshots below, try to open image as whole in new tab if you find something interesting. I could not crop each snapshot separately. For better clarity you can right click and open image in new tab. The three videos are as follows:
- The Biggest Mystery in the History of the Universe | Joana Xavier on the Origin of Life
- From Rebellious Teenager to Young Adult: Intelligent Design Grows Up
- Physics of Emergent Behaviour III: from origin of life to multicellularity, 2nd July 2021 (part 1)
Slide 2: Life? It highlights the inspiring nature of scientific discovery and suggesting a link between the natural world, represented by the Beryl crystal structure, and the discussion on life and its origins, as hinted by the question mark after "Life?". It also emphasize the wonder and fascination that scientists have for the natural world, as expressed by Marie Curie's quote.
Slide 3: Prokaryotes, which are a type of simple, single-celled organisms that lack a true cell nucleus. The importance of prokaryotes as some of the simplest and most widespread life forms on Earth. The comparison with the number of stars in the universe serves to emphasize their prevalence and significance in the context of the presentation on the origins of life on Earth.
Super inspiring keynote by @joanarcxavier at #WIVACE2023 @CaFoscari on Autocatalysis and the origins of life on Earth (though I do not fully agree with her on chemistry 🦠 being the only possible substrate for life 🧬) #ArtificialLife #ALife 🤖 pic.twitter.com/vh3O5HQ2bP
— Stefano Nichele (@stenichele) September 6, 2023
Slide 4: Cellular complexity has been unknown, underappreciated, neglected. It 4 draws attention to the fact that the complexity of cellular structures, particularly in prokaryotic cells like mycoplasma, has often been overlooked or underestimated. It introduces a specific research effort by Martina Martian and her team, who have been working on building structural models of mycoplasma cells. This slide could serve to emphasize the need for a deeper understanding of cellular complexity in the context of the presentation on the origins of life on Earth.
Slide 5: Universally essential elements in prokaryotes. A very nice interactive demo for this slide is availabe here which describes the cell structure in details. I am sorry I am not a biologist by education, so skipping this explanation. However really liked the web demo here Molecular Machinery: A tour of the protein data bank (Must try! A nice interactive website).
Slide 6: Metabolism. Slide 6 provides a comprehensive overview of metabolism in prokaryotic cells, highlighting its essential role in homeostasis, growth, and replication. It also emphasizes the efficiency of these organisms' genomes, the complexity of metabolic reactions, and the core essential components that underpin prokaryotic life. The notion of collective autocatalysis suggests a self-sustaining and interconnected nature of these processes within the cell. Persoanlly, I believe along with Homeostasis (which is a fundamental biological concept that refers to the ability of an organism or a system to maintain a stable internal environment despite external changes. It is a dynamic equilibrium that allows living organisms to function optimally by regulating various physiological variables within a narrow range. Homeostasis is crucial for the survival and proper functioning of organisms) Epigenetic Autonomy and Epigenetic Adaptation both are very important for Artificial Life. But anyways, Dr. Joana was not talking about ALife though in the slides. In fact, she was taking quite a stand on the ALife right now that it can only be improved or sustained if it can fit on homeostasis.
Slide 7: Finding universally essential cofactors for prokaryotes. (Lot of Biology, phew 😮💨) Skipping the details.
Slide 8: Some super interesting resulkts. (But again, Lot of Biology, phew 😮💨) Skipping the details.
Slide 9: Origins of Life = Origin of (proto-)cells. Reference: Dr. Joana Publications. BioChemistry = GeoChemistry. Slide 9 highlights the integral relationship between the origins of life and the concept of protocells, as well as the interplay between the chemical processes of life (biochemistry) and those of the Earth's environment (geochemistry).
Rest of the slides were highly centred to theoretical claims from past about homeostasis and autopoeisis. Please have a look for these slides in below caraousel (slides snapshots are already in sequence)
Finally we went for SPRITZ - Venice (Santa Croce) and then finally closed the Day 1 with dinner together ❤️.

Day 2 - 7th September 2023
There was no keynote and hence both the sessions of morning and afternoon were paper-presentation sessions. Out of which I liked some of the papers very interesting and hence I have prepared a short summary of these papers. I present following papers / sessions summaries in this section.- M4 Morning session 4: (25 min) Lorenzo Del Moro, Maurizio Magarini and Pasquale Stano On the Evaluation of Observed Semantic Information in Synthetic Cells - Click here for Summary!
- A3 Afternoon Session 3: (25 min) Luisa Damiano and Pasquale Stano “General Lines, Routes and Perspectives of Wetware Embodied AI. From its Organizational Bases to a Glimpse on Social Chemical Robotics” - Click here for Summary!
- A3 Afternoon Session 3: (25 min) Michele Braccini, Paolo Baldini and Andrea Roli An investigation of graceful degradation in Boolean network robots subject to online adaptation - Click here for Summary!
- A4 Afternoon Session 4: (25 min) Jørgen Jensen Farner, Ola Huse Ramstad, Stefano Nichele and Kristine Heiney Beyond weight plasticity: Local learning with propagation delays in spiking neural networks - Click here for Summary!
Graceful degradation of adaptive RBN robots by Paolo Baldini #WIVACE2023 pic.twitter.com/gio3XY1wfX
— Stefano Nichele (@stenichele) September 7, 2023
We have a double feature back to back from @stenichele lab, with Kristine Heiney talking about how to learn propagation delays in spiking neural networks. #wivace2023 pic.twitter.com/zWPrZIDULa
— JJ Merelo (@jjmerelo) September 7, 2023
Right now, @s4nyam presenting in #Wivace2034 his work on the AEVoT algorithm pic.twitter.com/ujUUfxadTe
— JJ Merelo (@jjmerelo) September 7, 2023
Evolving complexity in Lenia @s4nyam #WIVACE2023 @hiofnorge pic.twitter.com/lV5jkC3KXN
— Stefano Nichele (@stenichele) September 7, 2023
Local learning with propagation delays in SNNs @KrisHeiney #WIVACE2023 @OsloMet @hiofnorge pic.twitter.com/b1mnixnnww
— Stefano Nichele (@stenichele) September 7, 2023
Learning about Venice, gondolas, energy consumption of evolutionary algorithms & that Java is here to stay by @jjmerelo 😎 pic.twitter.com/WyNi8fE3GH
— Stefano Nichele (@stenichele) September 7, 2023
Day 3 - 8th September 2023
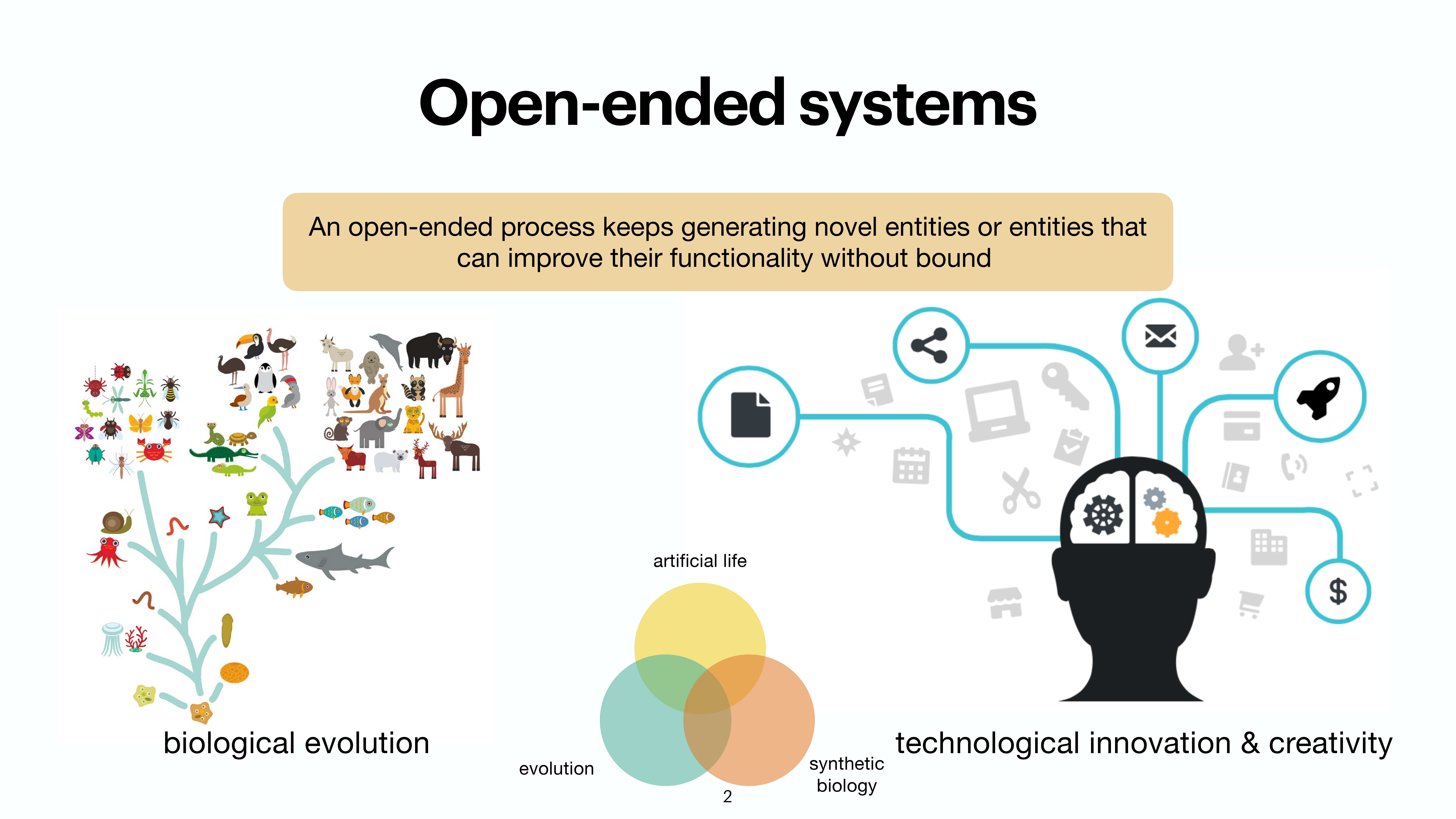
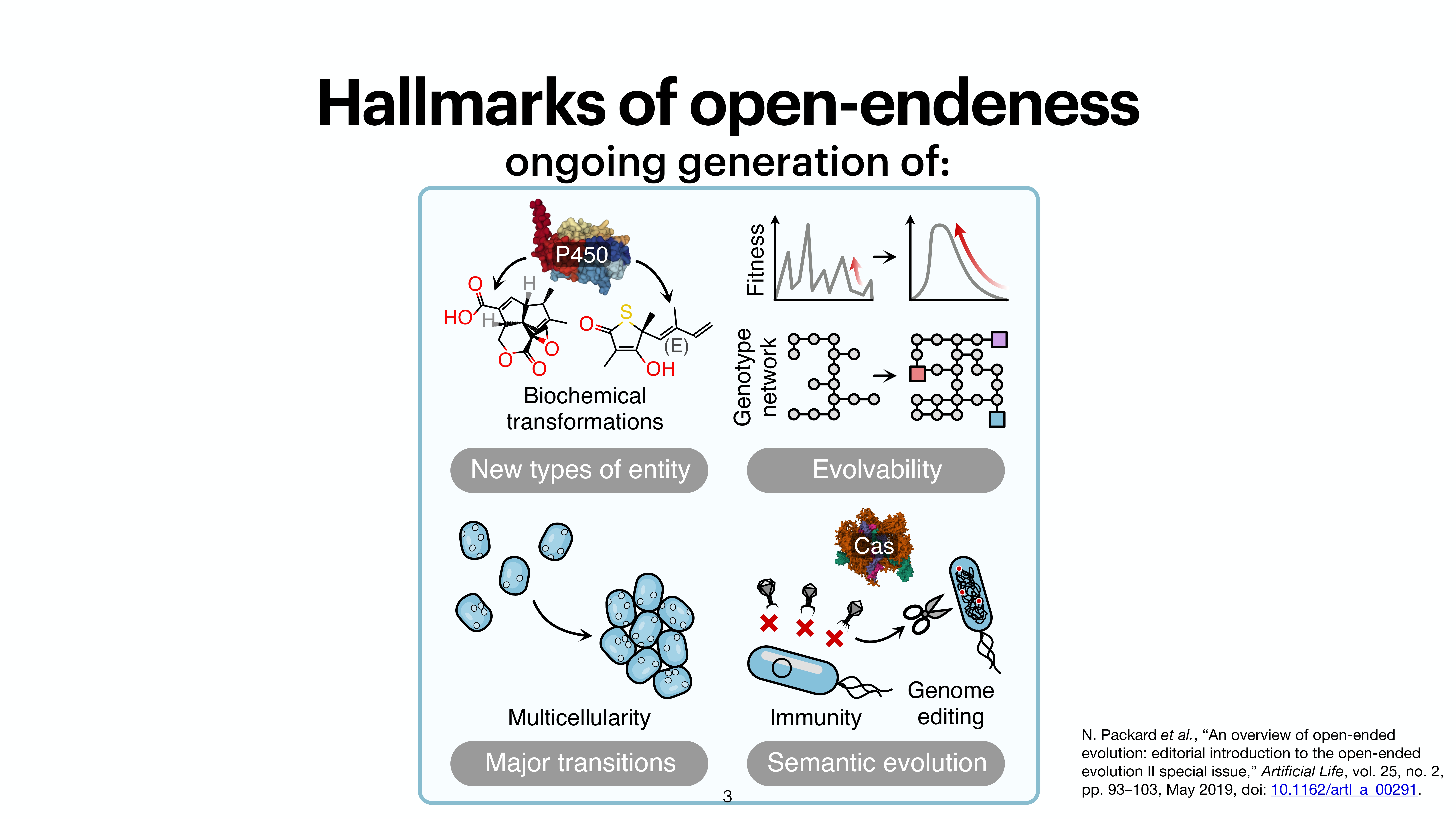
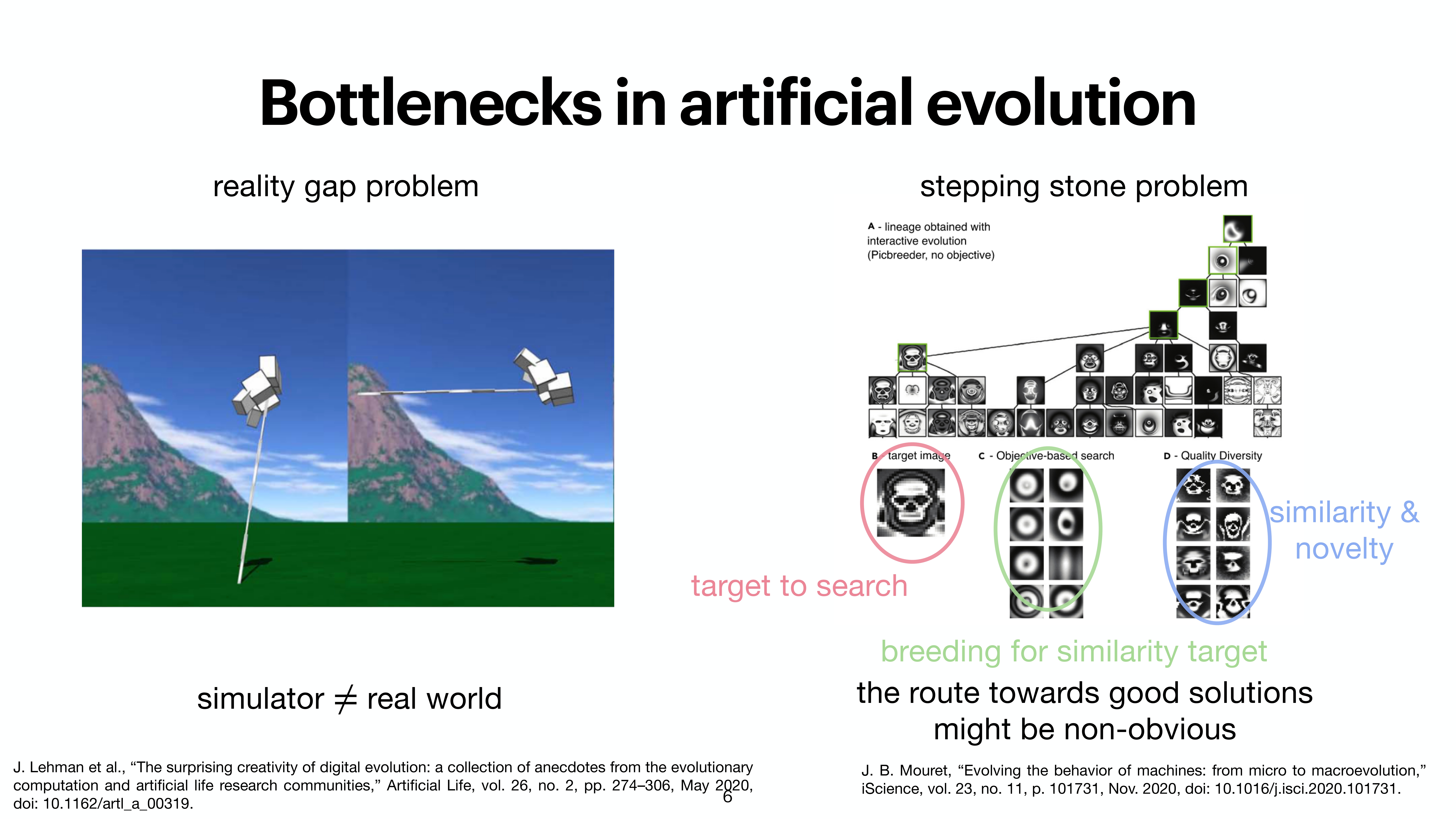
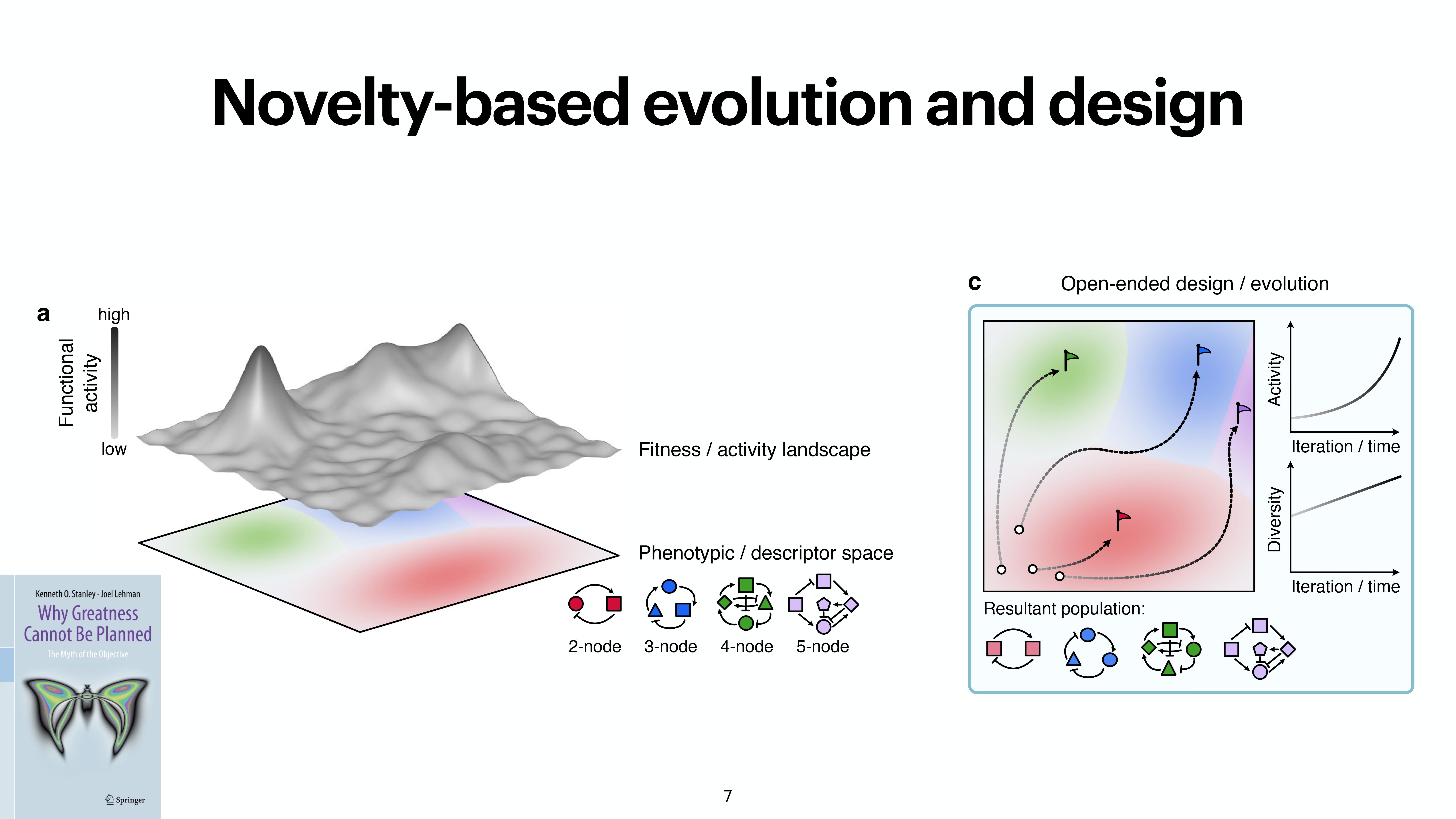
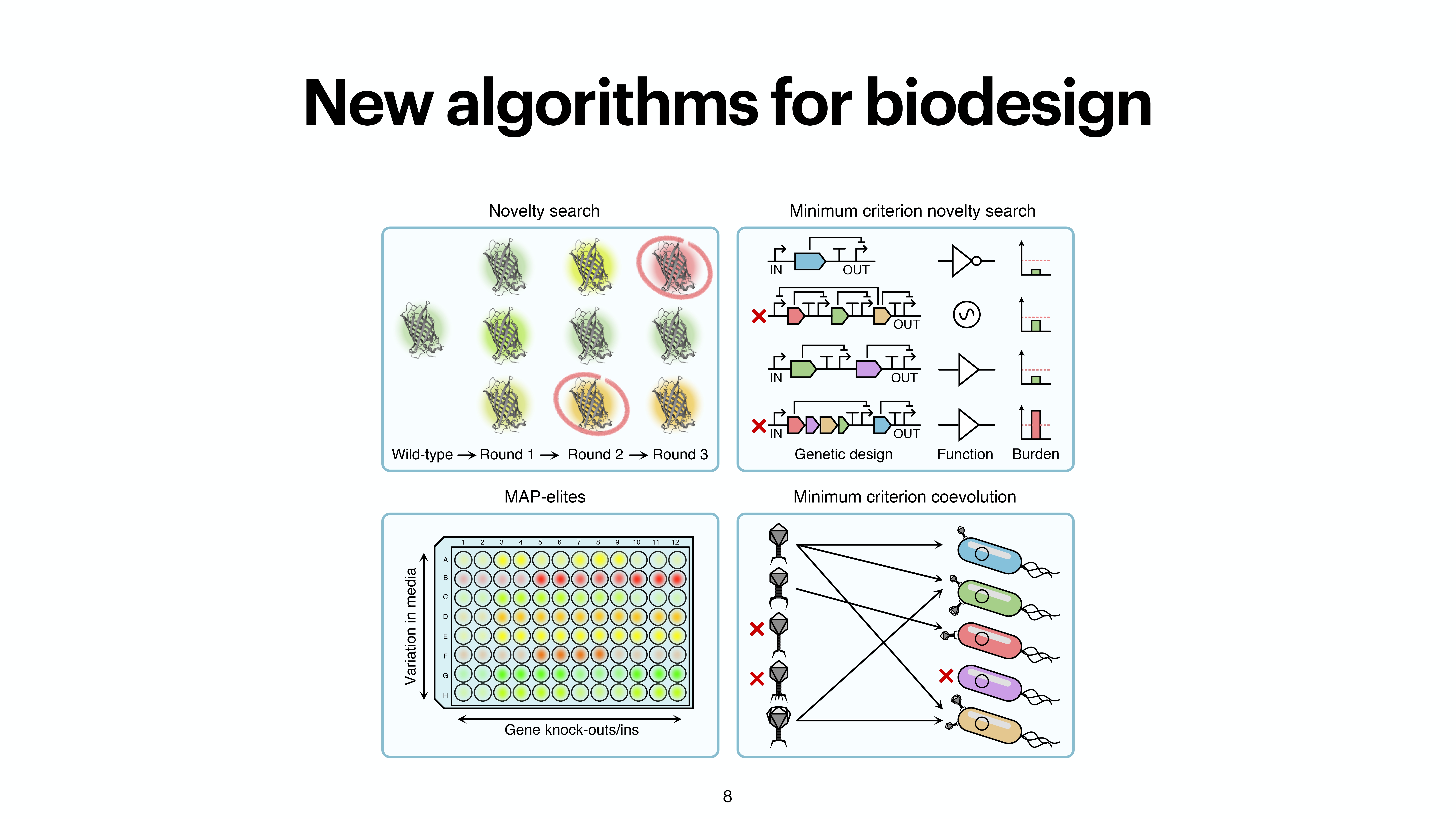
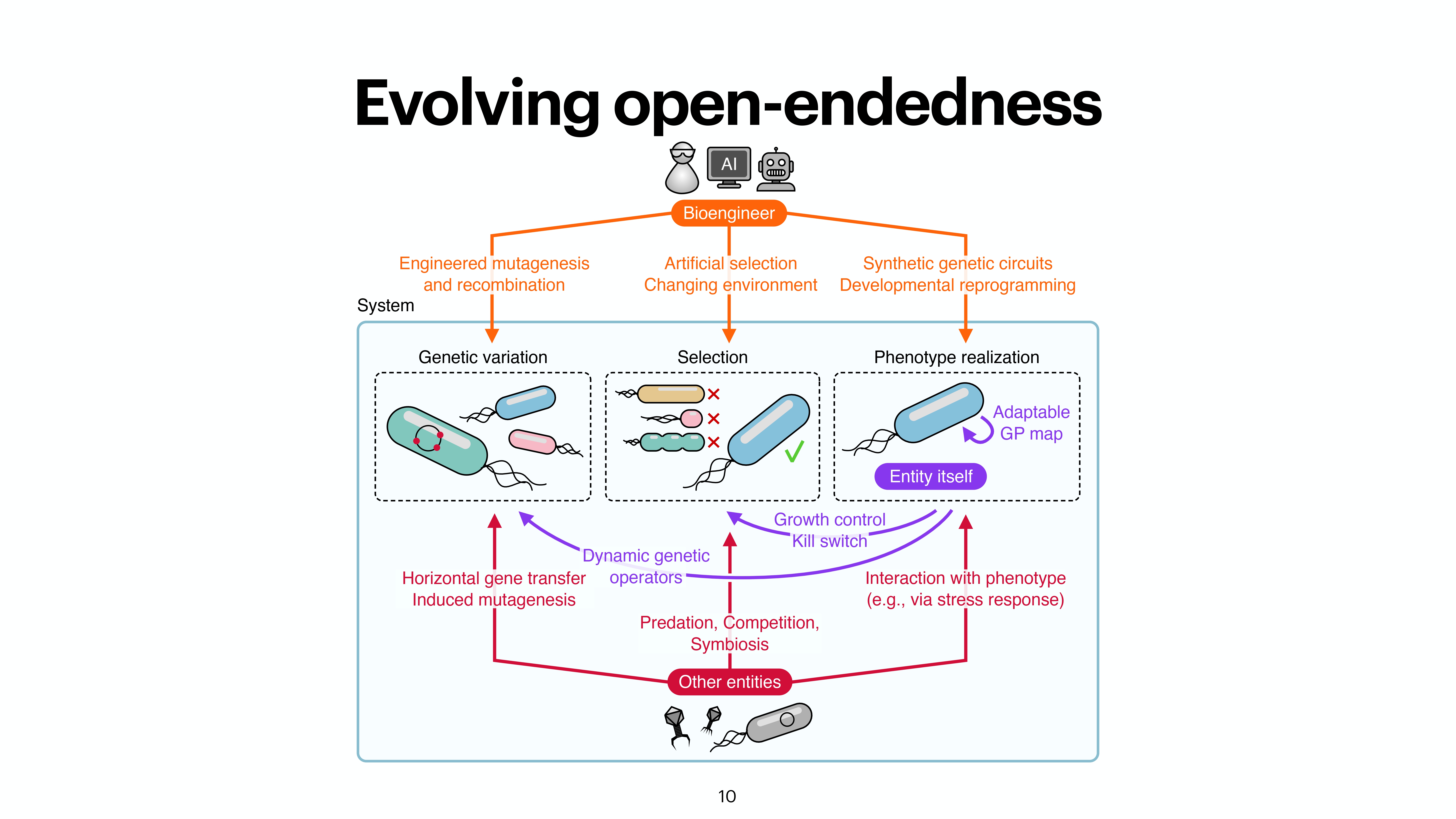
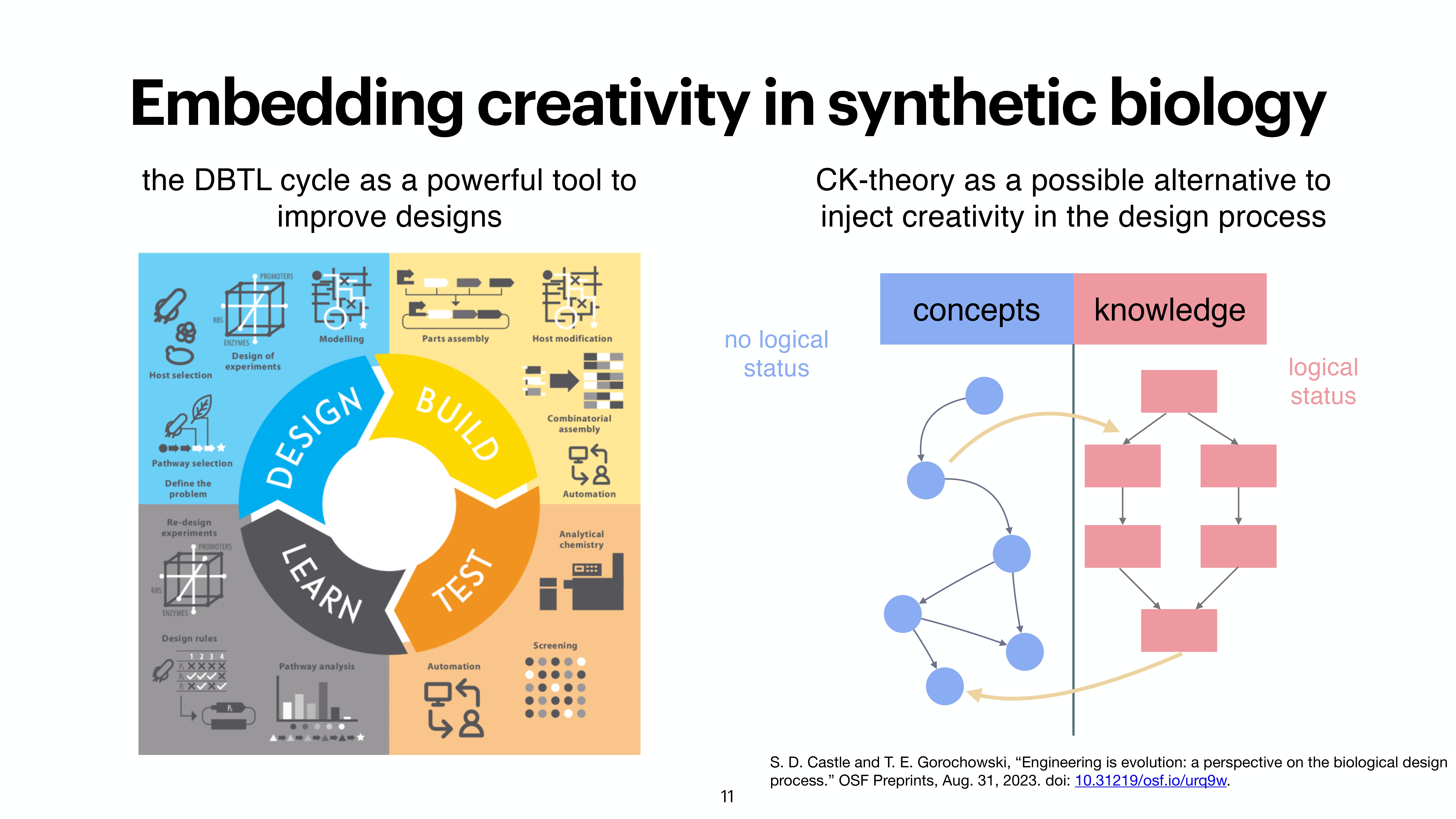
At the last day of the conference, there were two keynotes and intersting 20 minute talks. While the kenotes were very interesting on the topics of protein folding by Michele Vendruscolo and another interesting keynote from Wolfgang Banzhaf, my interest got more inclined towards talk by Michiel Stock, where he explained How to make everything: open-endedness in synthetic biology. Upon asking, prof provided the slides of the conference presentation by Michiel Stock, however, first lets check out some tweets from day 3.
Wolfgang Banzhaf @banzhaf_lab giving a keynote on Artificial Chemistries #WIVACE2023 @CaFoscari pic.twitter.com/rjVtLEFE6N
— Stefano Nichele (@stenichele) September 8, 2023
A nice work from @Michielstock on “How to make everything: open-endedness in synthetic biology” at #WIVACE2023 @CaFoscari 👏 pic.twitter.com/bLmWjTUWvN
— Sanyam Jain (@s4nyam) September 8, 2023
Prof Andrea nice talk on “The Hiatus Between Organism and Machine Evolution” 👏 at #WIVACE2023 pic.twitter.com/prLEko0XlD
— Sanyam Jain (@s4nyam) September 8, 2023
Slides from the presentation from Michiel Stock